6 Soil physical and hydrological properties
You are reading the work-in-progress An Open Compendium of Soil Sample and Soil Profile Datasets. This chapter is currently draft version, a peer-review publication is pending. You can find the polished first edition at https://opengeohub.github.io/SoilSamples/.
Last update: 2023-05-10
6.1 Overview
This section describes import steps used to produce a global compilation of soil laboratory data with physicals and hydraulic soil properties that can be then used for predictive soil mapping / modeling at global and regional scales.
Read more about computing with soil hydraulic / physical properties in R:
- Gupta, S., Hengl, T., Lehmann, P., Bonetti, S., and Or, D. SoilKsatDB: global soil saturated hydraulic conductivity measurements for geoscience applications. Earth Syst. Sci. Data Discuss., https://doi.org/10.5194/essd-2020-149, in review, 2021.
- de Sousa, D. F., Rodrigues, S., de Lima, H. V., & Chagas, L. T. (2020). R software packages as a tool for evaluating soil physical and hydraulic properties. Computers and Electronics in Agriculture, 168, 105077.
6.2  Specifications
Specifications
6.2.0.2 Target variables:
site.names = c("site_key", "usiteid", "site_obsdate", "longitude_decimal_degrees", "latitude_decimal_degrees", "location_accuracy_min", "location_accuracy_max")
hor.names = c("labsampnum","site_key","layer_sequence","hzn_top","hzn_bot","hzn_desgn","db_13b", "db_od", "COLEws", "w6clod", "w10cld", "w3cld", "w15l2", "w15bfm", "adod", "wrd_ws13", "cec7_cly", "w15cly", "tex_psda", "clay_tot_psa", "silt_tot_psa", "sand_tot_psa", "oc", "ph_kcl", "ph_h2o", "cec_sum", "cec_nh4", "wpg2", "ksat_lab", "ksat_field")
## target structure:
col.names = c("site_key", "usiteid", "site_obsdate", "longitude_decimal_degrees", "latitude_decimal_degrees", "location_accuracy_min", "location_accuracy_max", "labsampnum", "layer_sequence", "hzn_top", "hzn_bot", "hzn_desgn", "db_13b", "db_od", "COLEws", "w6clod", "w10cld", "w3cld", "w15l2", "w15bfm", "adod", "wrd_ws13", "cec7_cly", "w15cly", "tex_psda", "clay_tot_psa", "silt_tot_psa", "sand_tot_psa", "oc", "ph_kcl", "ph_h2o", "cec_sum", "cec_nh4", "wpg2", "ksat_lab", "ksat_field", "source_db", "confidence_degree", "project_url", "citation_url")-
db_13b: Bulk density (33kPa) in g/cm3 for <2mm soil fraction, -
db: Bulk density (unknown method) in g/cm3 for <2mm soil fraction, -
COLEws: Coefficient of Linear Extensibility (COLE) whole soil in ratio for <2mm soil fraction, -
w6clod: Water Content 6 kPa <2mm in % wt for <2mm soil fraction, -
w10cld: Water Content 10 kPa <2mm in % wt for <2mm soil fraction, -
w3cld: Water Content 33 kPa <2mm in % vol for <2mm soil fraction (Field Capacity), -
w15l2: Water Content 1500 kPa <2mm in % vol for <2mm soil fraction (Permanent Wilting Point), -
w15bfm: Water Content 1500 kPa moist <2mm in % wt for <2mm soil fraction, -
adod: Air-Dry/Oven-Dry in ratio for <2mm soil fraction, -
wrd_ws13: Water Retention Difference whole soil, 1500-kPa suction and an upper limit of usually 33-kPa in cm3 / cm-3 for <2mm soil fraction, -
cec7_cly: CEC-7/Clay ratio in ratio for <2mm soil fraction, -
w15cly: CEC/Clay ratio at 1500 kPa in ratio for <2mm soil fraction, -
tex_psda: Texture Determined, PSDA in factor for <2mm soil fraction, -
clay_tot_psa: Total Clay, <0.002 mm (<2 µm) in % wt for <2mm soil fraction, -
silt_tot_psa: Total Silt, 0.002-0.05 mm in % wt for <2mm soil fraction, -
sand_tot_psa: Total Sand, 0.05-2.0 mm in % wt for <2mm soil fraction, -
wpg2: Coarse fragments >2-mm weight fraction in % wt for <2mm soil fraction, -
hzn_top: The top (upper) depth of the layer in centimeters. in cm for <2mm soil fraction, -
hzn_bot: The bottom (lower) depth of the layer in centimeters. in cm for <2mm soil fraction, -
oc_v: Organic carbon (unknown method) in % wt for <2mm soil fraction, -
ph_kcl: pH, 1N KCl in ratio for <2mm soil fraction, -
ph_h2o_v: pH in water (unknown method) for <2mm soil fraction, -
cec_sum: Sum of Cations (CEC-8.2) in cmol(+)/kg for <2mm soil fraction, -
cec_nh4: NH4OAc, pH 7 (CEC-7) in cmol(+)/kg for <2mm soil fraction, -
ksat_field: Field-estimated Saturated Hydraulic Conductivity in cm/day for <2mm soil fraction, -
ksat_lab: Laboratory-estimated Saturated Hydraulic Conductivity in cm/day for <2mm soil fraction,
Same variable names have been adjusted (e.g. ph_h2o to ph_h2o_na) to include unknown method so that variables from different laboratory methods can be seamlessly merged.
Conversion between VWC and MWC is based on the formula (landon1991handbook?; benham1998field?; vanReeuwijk1993procedures?):
- VWC (%v/v) = MWC (% by weight ) * bulk density (kg/m3)
6.3  Data import
Data import
6.3.0.1 NCSS Characterization Database
- National Cooperative Soil Survey, (2020). National Cooperative Soil Survey Characterization Database. http://ncsslabdatamart.sc.egov.usda.gov/
if(!exists("hydrosprops.NCSS")){
ncss.site <- read.csv("/mnt/diskstation/data/Soil_points/INT/USDA_NCSS/NCSS_Site_Location.csv", stringsAsFactors = FALSE)
#str(ncss.site)
## Location accuracy unknown but we assume 100m
ncss.site$location_accuracy_max = NA
ncss.site$location_accuracy_min = 100
ncss.layer <- read.csv("/mnt/diskstation/data/Soil_points/INT/USDA_NCSS/NCSS_Layer.csv", stringsAsFactors = FALSE)
ncss.bdm <- read.csv("/mnt/diskstation/data/Soil_points/INT/USDA_NCSS/NCSS_Bulk_Density_and_Moisture.csv", stringsAsFactors = FALSE)
#summary(as.factor(ncss.bdm$prep_code))
ncss.bdm.0 <- ncss.bdm[ncss.bdm$prep_code=="S",]
#summary(ncss.bdm.0$db_od)
## 0 values --- error!
ncss.carb <- read.csv("/mnt/diskstation/data/Soil_points/INT/USDA_NCSS/NCSS_Carbon_and_Extractions.csv", stringsAsFactors = FALSE)
ncss.organic <- read.csv("/mnt/diskstation/data/Soil_points/INT/USDA_NCSS/NCSS_Organic.csv", stringsAsFactors = FALSE)
ncss.pH <- read.csv("/mnt/diskstation/data/Soil_points/INT/USDA_NCSS/NCSS_pH_and_Carbonates.csv", stringsAsFactors = FALSE)
#str(ncss.pH)
#summary(!is.na(ncss.pH$ph_h2o))
ncss.PSDA <- read.csv("/mnt/diskstation/data/Soil_points/INT/USDA_NCSS/NCSS_PSDA_and_Rock_Fragments.csv", stringsAsFactors = FALSE)
ncss.CEC <- read.csv("/mnt/diskstation/data/Soil_points/INT/USDA_NCSS/NCSS_CEC_and_Bases.csv")
ncss.horizons <- plyr::join_all(list(ncss.bdm.0, ncss.layer, ncss.carb, ncss.organic, ncss.pH, ncss.PSDA, ncss.CEC), type = "left", by="labsampnum")
#head(ncss.horizons)
nrow(ncss.horizons)
ncss.horizons$ksat_lab = NA; ncss.horizons$ksat_field = NA
hydrosprops.NCSS = plyr::join(ncss.site[,site.names], ncss.horizons[,hor.names], by="site_key")
## soil organic carbon:
#summary(!is.na(hydrosprops.NCSS$oc))
#summary(!is.na(hydrosprops.NCSS$ph_h2o))
#summary(!is.na(hydrosprops.NCSS$ph_kcl))
hydrosprops.NCSS$source_db = "USDA_NCSS"
#str(hydrosprops.NCSS)
#hist(hydrosprops.NCSS$w3cld[hydrosprops.NCSS$w3cld<150], breaks=45, col="gray")
## ERROR: MANY VALUES >100%
## fills in missing BD values using formula from Köchy, Hiederer, and Freibauer (2015)
db.f = ifelse(is.na(hydrosprops.NCSS$db_13b), -0.31*log(hydrosprops.NCSS$oc)+1.38, hydrosprops.NCSS$db_13b)
db.f[db.f<0.02 | db.f>2.87] = NA
## Convert to volumetric % to match most of world data sets:
hydrosprops.NCSS$w3cld = hydrosprops.NCSS$w3cld * db.f
hydrosprops.NCSS$w15l2 = hydrosprops.NCSS$w15l2 * db.f
hydrosprops.NCSS$w10cld = hydrosprops.NCSS$w10cld * db.f
#summary(as.factor(hydrosprops.NCSS$tex_psda))
## texture classes need to be cleaned up!
## check WRC values for sandy soils
#hydrosprops.NCSS[which(!is.na(hydrosprops.NCSS$w3cld) & hydrosprops.NCSS$sand_tot_psa>95)[1:10],]
## check WRC values for ORGANIC soils
#hydrosprops.NCSS[which(!is.na(hydrosprops.NCSS$w3cld) & hydrosprops.NCSS$oc>12)[1:10],]
## w3cld > 100?
hydrosprops.NCSS$confidence_degree = 1
hydrosprops.NCSS$project_url = "http://ncsslabdatamart.sc.egov.usda.gov/"
hydrosprops.NCSS$citation_url = "https://doi.org/10.2136/sssaj2016.11.0386n"
hydrosprops.NCSS = complete.vars(hydrosprops.NCSS)
saveRDS.gz(hydrosprops.NCSS, "/mnt/diskstation/data/Soil_points/INT/USDA_NCSS/hydrosprops.NCSS.rds")
}
dim(hydrosprops.NCSS)
#> [1] 113991 406.3.0.2 Africa soil profiles database
- Leenaars, J. G., Van OOstrum, A. J. M., & Ruiperez Gonzalez, M. (2014). Africa soil profiles database version 1.2. A compilation of georeferenced and standardized legacy soil profile data for Sub-Saharan Africa (with dataset). Wageningen: ISRIC Report 2014/01; 2014.
if(!exists("hydrosprops.AfSPDB")){
require(foreign)
afspdb.profiles <- read.dbf("/mnt/diskstation/data/Soil_points/AF/AfSIS_SPDB/AfSP012Qry_Profiles.dbf", as.is=TRUE)
## approximate location error
afspdb.profiles$location_accuracy_min = afspdb.profiles$XYAccur * 1e5
afspdb.profiles$location_accuracy_min = ifelse(afspdb.profiles$location_accuracy_min < 20, NA, afspdb.profiles$location_accuracy_min)
afspdb.profiles$location_accuracy_max = NA
afspdb.layers <- read.dbf("/mnt/diskstation/data/Soil_points/AF/AfSIS_SPDB/AfSP012Qry_Layers.dbf", as.is=TRUE)
## select columns of interest:
afspdb.s.lst <- c("ProfileID", "usiteid", "T_Year", "X_LonDD", "Y_LatDD", "location_accuracy_min", "location_accuracy_max")
## Convert to weight content
#summary(afspdb.layers$BlkDens)
## select layers
afspdb.h.lst <- c("LayerID", "ProfileID", "LayerNr", "UpDpth", "LowDpth", "HorDes", "db_13b", "BlkDens", "COLEws", "VMCpF18", "VMCpF20", "VMCpF25", "VMCpF42", "w15bfm", "adod", "wrd_ws13", "cec7_cly", "w15cly", "LabTxtr", "Clay", "Silt", "Sand", "OrgC", "PHKCl", "PHH2O", "CecSoil", "cec_nh4", "CfPc", "ksat_lab", "ksat_field")
## add missing columns
for(j in c("usiteid")){ afspdb.profiles[,j] = NA }
for(j in c("db_13b", "COLEws", "w15bfm", "adod", "wrd_ws13", "cec7_cly", "w15cly", "cec_nh4", "ksat_lab", "ksat_field")){ afspdb.layers[,j] = NA }
hydrosprops.AfSPDB = plyr::join(afspdb.profiles[,afspdb.s.lst], afspdb.layers[,afspdb.h.lst])
for(j in 1:ncol(hydrosprops.AfSPDB)){
if(is.numeric(hydrosprops.AfSPDB[,j])) { hydrosprops.AfSPDB[,j] <- ifelse(hydrosprops.AfSPDB[,j] < -200, NA, hydrosprops.AfSPDB[,j]) }
}
hydrosprops.AfSPDB$source_db = "AfSPDB"
hydrosprops.AfSPDB$confidence_degree = 5
hydrosprops.AfSPDB$OrgC = hydrosprops.AfSPDB$OrgC/10
#summary(hydrosprops.AfSPDB$OrgC)
hydrosprops.AfSPDB$project_url = "https://www.isric.org/projects/africa-soil-profiles-database-afsp"
hydrosprops.AfSPDB$citation_url = "https://www.isric.org/sites/default/files/isric_report_2014_01.pdf"
hydrosprops.AfSPDB = complete.vars(hydrosprops.AfSPDB, sel = c("VMCpF25", "VMCpF42"), coords = c("X_LonDD", "Y_LatDD"))
saveRDS.gz(hydrosprops.AfSPDB, "/mnt/diskstation/data/Soil_points/AF/AfSIS_SPDB/hydrosprops.AfSPDB.rds")
}
dim(hydrosprops.AfSPDB)
#> [1] 10720 406.3.0.3 ISRIC ISIS
- Batjes, N. H. (1995). A homogenized soil data file for global environmental research: A subset of FAO, ISRIC and NRCS profiles (Version 1.0) (No. 95/10b). ISRIC.
- Van de Ven, T., & Tempel, P. (1994). ISIS 4.0: ISRIC Soil Information System: User Manual. ISRIC.
if(!exists("hydrosprops.ISIS")){
isis.xy <- read.csv("/mnt/diskstation/data/Soil_points/INT/ISRIC_ISIS/Sites.csv", stringsAsFactors = FALSE)
#str(isis.xy)
isis.des <- read.csv("/mnt/diskstation/data/Soil_points/INT/ISRIC_ISIS/SitedescriptionResults.csv", stringsAsFactors = FALSE)
isis.site <- data.frame(site_key=isis.xy$Id, usiteid=paste(isis.xy$CountryISO, isis.xy$SiteNumber, sep=""))
id0.lst = c(236,235,224)
nm0.lst = c("longitude_decimal_degrees", "latitude_decimal_degrees", "site_obsdate")
isis.site.l = plyr::join_all(lapply(1:length(id0.lst), function(i){plyr::rename(subset(isis.des, ValueId==id0.lst[i])[,c("SampleId","Value")], replace=c("SampleId"="site_key", "Value"=paste(nm0.lst[i])))}), type = "full")
isis.site.df = join(isis.site, isis.site.l)
for(j in nm0.lst){ isis.site.df[,j] <- as.numeric(isis.site.df[,j]) }
isis.site.df[isis.site.df$usiteid=="CI2","latitude_decimal_degrees"] = 5.883333
#str(isis.site.df)
isis.site.df$location_accuracy_min = 100
isis.site.df$location_accuracy_max = NA
isis.smp <- read.csv("/mnt/diskstation/data/Soil_points/INT/ISRIC_ISIS/AnalyticalSamples.csv", stringsAsFactors = FALSE)
isis.ana <- read.csv("/mnt/diskstation/data/Soil_points/INT/ISRIC_ISIS/AnalyticalResults.csv", stringsAsFactors = FALSE)
#str(isis.ana)
isis.class <- read.csv("/mnt/diskstation/data/Soil_points/INT/ISRIC_ISIS/ClassificationResults.csv", stringsAsFactors = FALSE)
isis.hor <- data.frame(labsampnum=isis.smp$Id, hzn_top=isis.smp$Top, hzn_bot=isis.smp$Bottom, site_key=isis.smp$SiteId)
isis.hor$hzn_bot <- as.numeric(gsub(">", "", isis.hor$hzn_bot))
#str(isis.hor)
id.lst = c(1,2,22,4,28,31,32,14,34,38,39,42)
nm.lst = c("ph_h2o","ph_kcl","wpg2","oc","sand_tot_psa","silt_tot_psa","clay_tot_psa","cec_sum","db_od","w10cld","w3cld", "w15l2")
#str(as.numeric(isis.ana$Value[isis.ana$ValueId==38]))
isis.hor.l = plyr::join_all(lapply(1:length(id.lst), function(i){plyr::rename(subset(isis.ana, ValueId==id.lst[i])[,c("SampleId","Value")], replace=c("SampleId"="labsampnum", "Value"=paste(nm.lst[i])))}), type = "full")
#summary(as.numeric(isis.hor.l$w3cld))
isis.hor.df = join(isis.hor, isis.hor.l)
isis.hor.df = isis.hor.df[!duplicated(isis.hor.df$labsampnum),]
#summary(as.numeric(isis.hor.df$w3cld))
for(j in nm.lst){ isis.hor.df[,j] <- as.numeric(isis.hor.df[,j]) }
#str(isis.hor.df)
## add missing columns
for(j in c("layer_sequence", "hzn_desgn", "tex_psda", "COLEws", "w15bfm", "adod", "wrd_ws13", "cec7_cly", "w15cly", "cec_nh4", "db_13b", "w6clod", "ksat_lab", "ksat_field")){ isis.hor.df[,j] = NA }
which(!hor.names %in% names(isis.hor.df))
hydrosprops.ISIS <- join(isis.site.df[,site.names], isis.hor.df[,hor.names], type="left")
hydrosprops.ISIS$source_db = "ISRIC_ISIS"
hydrosprops.ISIS$confidence_degree = 1
hydrosprops.ISIS$project_url = "https://isis.isric.org"
hydrosprops.ISIS$citation_url = "https://www.isric.org/sites/default/files/isric_report_1995_10b.pdf"
hydrosprops.ISIS = complete.vars(hydrosprops.ISIS)
saveRDS.gz(hydrosprops.ISIS, "/mnt/diskstation/data/Soil_points/INT/ISRIC_ISIS/hydrosprops.ISIS.rds")
}
dim(hydrosprops.ISIS)
#> [1] 1176 406.3.0.4 ISRIC WISE
- Batjes, N.H. (2019). Harmonized soil profile data for applications at global and continental scales: updates to the WISE database. Soil Use and Management 5:124–127.
if(!exists("hydrosprops.WISE")){
wise.SITE <- read.csv("/mnt/diskstation/data/Soil_points/INT/ISRIC_WISE/WISE3_SITE.csv", stringsAsFactors=FALSE)
#summary(as.factor(wise.SITE$LONLAT_ACC))
wise.SITE$location_accuracy_min = ifelse(wise.SITE$LONLAT_ACC=="D", 1e5/2, ifelse(wise.SITE$LONLAT_ACC=="S", 30, ifelse(wise.SITE$LONLAT_ACC=="M", 1800/2, NA)))
wise.SITE$location_accuracy_max = NA
wise.HORIZON <- read.csv("/mnt/diskstation/data/Soil_points/INT/ISRIC_WISE/WISE3_HORIZON.csv")
wise.s.lst <- c("WISE3_id", "SOURCE_ID", "DATEYR", "LONDD", "LATDD", "location_accuracy_min", "location_accuracy_max")
## Volumetric values
#summary(wise.HORIZON$BULKDENS)
#summary(wise.HORIZON$VMC1)
wise.HORIZON$WISE3_id = wise.HORIZON$WISE3_ID
wise.h.lst <- c("labsampnum", "WISE3_id", "HONU", "TOPDEP", "BOTDEP", "DESIG", "db_13b", "BULKDENS", "COLEws", "w6clod", "VMC1", "VMC2", "VMC3", "w15bfm", "adod", "wrd_ws13", "cec7_cly", "w15cly", "tex_psda", "CLAY", "SILT", "SAND", "ORGC", "PHKCL", "PHH2O", "CECSOIL", "cec_nh4", "GRAVEL", "ksat_lab", "ksat_field")
## add missing columns
for(j in c("labsampnum", "db_13b", "COLEws", "w15bfm", "w6clod", "adod", "wrd_ws13", "cec7_cly", "w15cly", "tex_psda", "cec_nh4", "ksat_lab", "ksat_field")){ wise.HORIZON[,j] = NA }
hydrosprops.WISE = plyr::join(wise.SITE[,wise.s.lst], wise.HORIZON[,wise.h.lst])
for(j in 1:ncol(hydrosprops.WISE)){
if(is.numeric(hydrosprops.WISE[,j])) { hydrosprops.WISE[,j] <- ifelse(hydrosprops.WISE[,j] < -200, NA, hydrosprops.WISE[,j]) }
}
hydrosprops.WISE$ORGC = hydrosprops.WISE$ORGC/10
hydrosprops.WISE$source_db = "ISRIC_WISE"
hydrosprops.WISE$project_url = "https://isric.org"
hydrosprops.WISE$citation_url = "http://dx.doi.org/10.1111/j.1475-2743.2009.00202.x"
hydrosprops.WISE <- complete.vars(hydrosprops.WISE, sel=c("VMC2", "VMC3"), coords = c("LONDD", "LATDD"))
hydrosprops.WISE$confidence_degree = 5
#summary(hydrosprops.WISE$VMC3)
saveRDS.gz(hydrosprops.WISE, "/mnt/diskstation/data/Soil_points/INT/ISRIC_WISE/hydrosprops.WISE.rds")
}
dim(hydrosprops.WISE)
#> [1] 1325 406.3.0.5 Fine Root Ecology Database (FRED)
- Iversen CM, Powell AS, McCormack ML, Blackwood CB, Freschet GT, Kattge J, Roumet C, Stover DB, Soudzilovskaia NA, Valverde-Barrantes OJ, van Bodegom PM, Violle C. 2018. Fine-Root Ecology Database (FRED): A Global Collection of Root Trait Data with Coincident Site, Vegetation, Edaphic, and Climatic Data, Version 2. Oak Ridge National Laboratory, TES SFA, U.S. Department of Energy, Oak Ridge, Tennessee, U.S.A. Access on-line at: https://doi.org/10.25581/ornlsfa.012/1417481.
if(!exists("hydrosprops.FRED")){
fred = read.csv("/mnt/diskstation/data/Soil_points/INT/FRED/FRED2_20180518.csv", skip = 5, header=FALSE)
names(fred) = names(read.csv("/mnt/diskstation/data/Soil_points/INT/FRED/FRED2_20180518.csv", nrows=1, header=TRUE))
fred.h.lst = c("Notes_Row.ID", "Data.source_DOI", "site_obsdate", "longitude_decimal_degrees", "latitude_decimal_degrees", "location_accuracy_min", "location_accuracy_max", "labsampnum", "layer_sequence", "hzn_top", "hzn_bot", "Soil.horizon", "db_13b", "Soil.bulk.density", "COLEws", "w6clod", "w10cld", "Soil.water_Volumetric.content", "w15l2", "w15bfm", "adod", "wrd_ws13", "cec7_cly", "w15cly", "Soil.texture", "Soil.texture_Fraction.clay", "Soil.texture_Fraction.silt", "Soil.texture_Fraction.sand", "Soil.organic.C.content", "ph_kcl", "Soil.pH_Water", "Soil.cation.exchange.capacity..CEC.", "cec_nh4", "wpg2", "ksat_lab", "ksat_field")
#summary(fred$Soil.water_Volumetric.content)
#summary(fred$Soil.water_Storage.capacity)
fred$site_obsdate = rowMeans(fred[,c("Sample.collection_Year.ending.collection", "Sample.collection_Year.beginning.collection")], na.rm=TRUE)
#summary(fred$site_obsdate)
fred$longitude_decimal_degrees = ifelse(is.na(fred$Longitude), fred$Longitude_Estimated, fred$Longitude)
fred$latitude_decimal_degrees = ifelse(is.na(fred$Latitude), fred$Latitude_Estimated, fred$Latitude)
#summary(as.factor(fred$Soil.horizon))
fred$hzn_bot = ifelse(is.na(fred$Soil.depth_Lower.sampling.depth), fred$Soil.depth - 5, fred$Soil.depth_Lower.sampling.depth)
fred$hzn_top = ifelse(is.na(fred$Soil.depth_Upper.sampling.depth), fred$Soil.depth + 5, fred$Soil.depth_Upper.sampling.depth)
x.na = fred.h.lst[which(!fred.h.lst %in% names(fred))]
if(length(x.na)>0){ for(i in x.na){ fred[,i] = NA } }
hydrosprops.FRED = fred[,fred.h.lst]
#plot(hydrosprops.FRED[,4:5])
hydrosprops.FRED$source_db = "FRED"
hydrosprops.FRED$confidence_degree = 5
hydrosprops.FRED$project_url = "https://roots.ornl.gov/"
hydrosprops.FRED$citation_url = "https://doi.org/10.25581/ornlsfa.012/1417481"
hydrosprops.FRED = complete.vars(hydrosprops.FRED, sel = c("Soil.water_Volumetric.content", "Soil.texture_Fraction.clay"))
saveRDS.gz(hydrosprops.FRED, "/mnt/diskstation/data/Soil_points/INT/FRED/hydrosprops.FRED.rds")
}
dim(hydrosprops.FRED)
#> [1] 3761 406.3.0.6 EGRPR
if(!exists("hydrosprops.EGRPR")){
russ.HOR = read.csv("/mnt/diskstation/data/Soil_points/Russia/EGRPR/Russia_EGRPR_soil_pedons.csv")
russ.HOR$SOURCEID = paste(russ.HOR$CardID, russ.HOR$SOIL_ID, sep="_")
russ.HOR$SNDPPT <- russ.HOR$TEXTSAF + russ.HOR$TEXSCM
russ.HOR$SLTPPT <- russ.HOR$TEXTSIC + russ.HOR$TEXTSIM + 0.8 * russ.HOR$TEXTSIF
russ.HOR$CLYPPT <- russ.HOR$TEXTCL + 0.2 * russ.HOR$TEXTSIF
## Correct texture fractions:
sumTex <- rowSums(russ.HOR[,c("SLTPPT","CLYPPT","SNDPPT")])
russ.HOR$SNDPPT <- russ.HOR$SNDPPT / ((sumTex - russ.HOR$CLYPPT) /(100 - russ.HOR$CLYPPT))
russ.HOR$SLTPPT <- russ.HOR$SLTPPT / ((sumTex - russ.HOR$CLYPPT) /(100 - russ.HOR$CLYPPT))
russ.HOR$oc <- russ.HOR$ORGMAT/1.724
## add missing columns
for(j in c("site_obsdate", "location_accuracy_min", "location_accuracy_max", "labsampnum", "db_13b", "COLEws", "w15bfm", "w6clod", "adod", "wrd_ws13", "cec7_cly", "w15cly", "tex_psda", "cec_nh4", "wpg2", "ksat_lab", "ksat_field")){ russ.HOR[,j] = NA }
russ.sel.h = c("SOURCEID", "SOIL_ID", "site_obsdate", "LONG", "LAT", "location_accuracy_min", "location_accuracy_max", "labsampnum", "HORNMB", "HORTOP", "HORBOT", "HISMMN", "db_13b", "DVOL", "COLEws", "w6clod", "WR10", "WR33", "WR1500", "w15bfm", "adod", "wrd_ws13", "cec7_cly", "w15cly", "tex_psda", "CLYPPT", "SLTPPT", "SNDPPT", "oc", "PHSLT", "PHH2O", "CECST", "cec_nh4", "wpg2","ksat_lab", "ksat_field")
hydrosprops.EGRPR = russ.HOR[,russ.sel.h]
hydrosprops.EGRPR$source_db = "Russia_EGRPR"
hydrosprops.EGRPR$confidence_degree = 2
hydrosprops.EGRPR$project_url = "http://egrpr.esoil.ru/"
hydrosprops.EGRPR$citation_url = "https://doi.org/10.19047/0136-1694-2016-86-115-123"
hydrosprops.EGRPR <- complete.vars(hydrosprops.EGRPR, sel=c("WR33", "WR1500"), coords = c("LONG", "LAT"))
#summary(hydrosprops.EGRPR$WR1500)
saveRDS.gz(hydrosprops.EGRPR, "/mnt/diskstation/data/Soil_points/Russia/EGRPR/hydrosprops.EGRPR.rds")
}
dim(hydrosprops.EGRPR)
#> [1] 1138 406.3.0.7 SPADE-2
- Hannam J.A., Hollis, J.M., Jones, R.J.A., Bellamy, P.H., Hayes, S.E., Holden, A., Van Liedekerke, M.H. and Montanarella, L. (2009). SPADE-2: The soil profile analytical database for Europe, Version 2.0 Beta Version March 2009. Unpublished Report, 27pp.
- Kristensen, J. A., Balstrøm, T., Jones, R. J. A., Jones, A., Montanarella, L., Panagos, P., and Breuning-Madsen, H.: Development of a harmonised soil profile analytical database for Europe: a resource for supporting regional soil management, SOIL, 5, 289–301, https://doi.org/10.5194/soil-5-289-2019, 2019.
if(!exists("hydrosprops.SPADE2")){
spade.PLOT <- read.csv("/mnt/diskstation/data/Soil_points/EU/SPADE/DAT_PLOT.csv")
#str(spade.PLOT)
spade.HOR <- read.csv("/mnt/diskstation/data/Soil_points/EU/SPADE/DAT_HOR.csv")
spade.PLOT = spade.PLOT[!spade.PLOT$LON_COOR_V>180 & spade.PLOT$LAT_COOR_V>20,]
#plot(spade.PLOT[,c("LON_COOR_V","LAT_COOR_V")])
spade.PLOT$location_accuracy_min = 100
spade.PLOT$location_accuracy_max = NA
#site.names = c("site_key", "usiteid", "site_obsdate", "longitude_decimal_degrees", "latitude_decimal_degrees")
spade.PLOT$ProfileID = paste(spade.PLOT$CNTY_C, spade.PLOT$PLOT_ID, sep="_")
spade.PLOT$T_Year = 2009
spade.s.lst <- c("PLOT_ID", "ProfileID", "T_Year", "LON_COOR_V", "LAT_COOR_V", "location_accuracy_min", "location_accuracy_max")
## standardize:
spade.HOR$SLTPPT <- spade.HOR$SILT1_V + spade.HOR$SILT2_V
spade.HOR$SNDPPT <- spade.HOR$SAND1_V + spade.HOR$SAND2_V + spade.HOR$SAND3_V
spade.HOR$PHIKCL <- NA
spade.HOR$PHIKCL[which(spade.HOR$PH_M %in% "A14")] <- spade.HOR$PH_V[which(spade.HOR$PH_M %in% "A14")]
spade.HOR$PHIHO5 <- NA
spade.HOR$PHIHO5[which(spade.HOR$PH_M %in% "A12")] <- spade.HOR$PH_V[which(spade.HOR$PH_M %in% "A12")]
#summary(spade.HOR$BD_V)
for(j in c("site_obsdate", "layer_sequence", "db_13b", "COLEws", "w15bfm", "w6clod", "w10cld", "adod", "wrd_ws13", "w15bfm", "cec7_cly", "w15cly", "tex_psda", "cec_nh4", "ksat_lab", "ksat_field")){ spade.HOR[,j] = NA }
spade.h.lst = c("HOR_ID","PLOT_ID","layer_sequence","HOR_BEG_V","HOR_END_V","HOR_NAME","db_13b", "BD_V", "COLEws", "w6clod", "w10cld", "WCFC_V", "WC4_V", "w15bfm", "adod", "wrd_ws13", "cec7_cly", "w15cly", "tex_psda", "CLAY_V", "SLTPPT", "SNDPPT", "OC_V", "PHIKCL", "PHIHO5", "CEC_V", "cec_nh4", "GRAV_C", "ksat_lab", "ksat_field")
hydrosprops.SPADE2 = plyr::join(spade.PLOT[,spade.s.lst], spade.HOR[,spade.h.lst])
hydrosprops.SPADE2$source_db = "SPADE2"
hydrosprops.SPADE2$confidence_degree = 15
hydrosprops.SPADE2$project_url = "https://esdac.jrc.ec.europa.eu/content/soil-profile-analytical-database-2"
hydrosprops.SPADE2$citation_url = "https://doi.org/10.1016/j.landusepol.2011.07.003"
hydrosprops.SPADE2 <- complete.vars(hydrosprops.SPADE2, sel=c("WCFC_V", "WC4_V"), coords = c("LON_COOR_V","LAT_COOR_V"))
#summary(hydrosprops.SPADE2$WC4_V)
#summary(is.na(hydrosprops.SPADE2$WC4_V))
#hist(hydrosprops.SPADE2$WC4_V, breaks=45, col="gray")
saveRDS.gz(hydrosprops.SPADE2, "/mnt/diskstation/data/Soil_points/EU/SPADE/hydrosprops.SPADE2.rds")
}
dim(hydrosprops.SPADE2)
#> [1] 1182 406.3.0.8 Canada National Pedon Database
if(!exists("hydrosprops.NPDB")){
NPDB.nm = c("NPDB_V2_sum_source_info.csv","NPDB_V2_sum_chemical.csv", "NPDB_V2_sum_horizons_raw.csv", "NPDB_V2_sum_physical.csv")
NPDB.HOR = plyr::join_all(lapply(paste0("/mnt/diskstation/data/Soil_points/Canada/NPDB/", NPDB.nm), read.csv), type = "full")
#str(NPDB.HOR)
#summary(NPDB.HOR$BULK_DEN)
## 0 values -> ERROR!
## add missing columns
NPDB.HOR$HISMMN = paste0(NPDB.HOR$HZN_MAS, NPDB.HOR$HZN_SUF, NPDB.HOR$HZN_MOD)
for(j in c("usiteid", "location_accuracy_max", "layer_sequence", "labsampnum", "db_13b", "COLEws", "w15bfm", "w6clod", "w10cld", "adod", "wrd_ws13", "cec7_cly", "w15cly", "tex_psda", "cec_nh4", "ph_kcl", "ksat_lab", "ksat_field")){ NPDB.HOR[,j] = NA }
npdb.sel.h = c("PEDON_ID", "usiteid", "CAL_YEAR", "DD_LONG", "DD_LAT", "CONF_METRS", "location_accuracy_max", "labsampnum", "layer_sequence", "U_DEPTH", "L_DEPTH", "HISMMN", "db_13b", "BULK_DEN", "COLEws", "w6clod", "w10cld", "RETN_33KP", "RETN_1500K", "RETN_HYGR", "adod", "wrd_ws13", "cec7_cly", "w15cly", "tex_psda", "T_CLAY", "T_SILT", "T_SAND", "CARB_ORG", "ph_kcl", "PH_H2O", "CEC", "cec_nh4", "VC_SAND", "ksat_lab", "ksat_field")
hydrosprops.NPDB = NPDB.HOR[,npdb.sel.h]
hydrosprops.NPDB$source_db = "Canada_NPDB"
hydrosprops.NPDB$confidence_degree = 1
hydrosprops.NPDB$project_url = "https://open.canada.ca/data/en/"
hydrosprops.NPDB$citation_url = "https://open.canada.ca/data/en/dataset/6457fad6-b6f5-47a3-9bd1-ad14aea4b9e0"
hydrosprops.NPDB <- complete.vars(hydrosprops.NPDB, sel=c("RETN_33KP", "RETN_1500K"), coords = c("DD_LONG", "DD_LAT"))
saveRDS.gz(hydrosprops.NPDB, "/mnt/diskstation/data/Soil_points/Canada/NPDB/hydrosprops.NPDB.rds")
}
dim(hydrosprops.NPDB)
#> [1] 404 406.3.0.9 ETH imported data from literature
- Digitized soil hydraulic measurements from the literature by the ETH Soil and Terrestrial Environmental Physics.
if(!exists("hydrosprops.ETH")){
xlsxFile = list.files(pattern="Global_soil_water_tables.xlsx", full.names = TRUE, recursive = TRUE)
wb = openxlsx::getSheetNames(xlsxFile)
eth.tbl = plyr::rbind.fill(
openxlsx::read.xlsx(xlsxFile, sheet = "ETH_imported_literature"),
openxlsx::read.xlsx(xlsxFile, sheet = "ETH_imported_literature_more"),
openxlsx::read.xlsx(xlsxFile, sheet = "ETH_extra_data set"),
openxlsx::read.xlsx(xlsxFile, sheet = "Tibetan_plateau"),
openxlsx::read.xlsx(xlsxFile, sheet = "Belgium_Vereecken_data"),
openxlsx::read.xlsx(xlsxFile, sheet = "Australia_dataset"),
openxlsx::read.xlsx(xlsxFile, sheet = "Florida_Soils_Ksat"),
openxlsx::read.xlsx(xlsxFile, sheet = "China_dataset"),
openxlsx::read.xlsx(xlsxFile, sheet = "Sand_dunes_Siberia_database"),
openxlsx::read.xlsx(xlsxFile, sheet = "New_data_4_03")
)
#dim(eth.tbl)
#summary(as.factor(eth.tbl$reference_source))
## Data quality tables
lab.ql = openxlsx::read.xlsx(xlsxFile, sheet = "Quality_per_site_key")
lab.cd = plyr::join(eth.tbl["site_key"], lab.ql)$confidence_degree
eth.tbl$confidence_degree = ifelse(is.na(eth.tbl$confidence_degree), lab.cd, eth.tbl$confidence_degree)
#summary(as.factor(eth.tbl$confidence_degree))
## missing columns
for(j in c("usiteid", "labsampnum", "layer_sequence", "db_13b", "COLEws", "adod", "wrd_ws13", "w15bfm", "w15cly", "cec7_cly", "w6clod", "w10cld", "ph_kcl", "cec_sum", "cec_nh4", "wpg2", "project_url", "citation_url")){ eth.tbl[,j] = NA }
hydrosprops.ETH = eth.tbl[,col.names]
col.names[which(!col.names %in% names(eth.tbl))]
hydrosprops.ETH$project_url = "https://step.ethz.ch/"
hydrosprops.ETH$citation_url = "https://doi.org/10.5194/essd-2020-149"
hydrosprops.ETH = complete.vars(hydrosprops.ETH)
#hist(hydrosprops.ETH$w15l2, breaks=45, col="gray")
#hist(log1p(hydrosprops.ETH$ksat_lab), breaks=45, col="gray")
saveRDS.gz(hydrosprops.ETH, "/mnt/diskstation/data/Soil_points/INT/hydrosprops.ETH.rds")
}
dim(hydrosprops.ETH)
#> [1] 9023 406.3.0.10 HYBRAS
- Ottoni, M. V., Ottoni Filho, T. B., Schaap, M. G., Lopes-Assad, M. L. R., & Rotunno Filho, O. C. (2018). Hydrophysical database for Brazilian soils (HYBRAS) and pedotransfer functions for water retention. Vadose Zone Journal, 17(1).
if(!exists("hydrosprops.HYBRAS")){
hybras.HOR = openxlsx::read.xlsx(xlsxFile, sheet = "HYBRAS.V1_integrated_tables_RAW")
#str(hybras.HOR)
## some points had only UTM coordinates and had to be manually coorected
## subset to unique values:
hybras.HOR = hybras.HOR[!duplicated(hybras.HOR$site_key),]
#summary(hybras.HOR$bulk_den)
#hist(hybras.HOR$ksat, breaks=35, col="grey")
## add missing columns
for(j in c("usiteid", "layer_sequence", "labsampnum", "db_13b", "COLEws", "w15bfm", "w6clod", "w10cld", "adod", "wrd_ws13", "cec7_cly", "w15cly", "cec_sum", "cec_nh4", "ph_kcl", "ph_h2o", "ksat_field", "uuid")){ hybras.HOR[,j] = NA }
hybras.HOR$w3cld = rowMeans(hybras.HOR[,c("theta20","theta50")], na.rm = TRUE)
hybras.sel.h = c("site_key", "usiteid", "year", "LongitudeOR", "LatitudeOR", "location_accuracy_min", "location_accuracy_max", "labsampnum", "layer_sequence", "top_depth", "bot_depth", "horizon", "db_13b", "bulk_den", "COLEws", "w6clod", "theta10", "w3cld", "theta15000", "satwat", "adod", "wrd_ws13", "cec7_cly", "w15cly", "tex_psda", "clay", "silt", "sand", "org_carb", "ph_kcl", "ph_h2o", "cec_sum", "cec_nh4", "vc_sand", "ksat", "ksat_field")
hydrosprops.HYBRAS = hybras.HOR[,hybras.sel.h]
hydrosprops.HYBRAS$source_db = "HYBRAS"
hydrosprops.HYBRAS$confidence_degree = 1
for(i in c("theta10", "w3cld", "theta15000", "satwat")){ hydrosprops.HYBRAS[,i] = hydrosprops.HYBRAS[,i]*100 }
#summary(hydrosprops.HYBRAS$theta10)
#summary(hydrosprops.HYBRAS$satwat)
#hist(hydrosprops.HYBRAS$theta10, breaks=45, col="gray")
#hist(log1p(hydrosprops.HYBRAS$ksat), breaks=45, col="gray")
#summary(!is.na(hydrosprops.HYBRAS$ksat))
hydrosprops.HYBRAS$project_url = "http://www.cprm.gov.br/en/Hydrology/Research-and-Innovation/HYBRAS-4208.html"
hydrosprops.HYBRAS$citation_url = "https://doi.org/10.2136/vzj2017.05.0095"
hydrosprops.HYBRAS <- complete.vars(hydrosprops.HYBRAS, sel=c("w3cld", "theta15000", "ksat", "ksat_field"), coords = c("LongitudeOR", "LatitudeOR"))
saveRDS.gz(hydrosprops.HYBRAS, "/mnt/diskstation/data/Soil_points/INT/HYBRAS/hydrosprops.HYBRAS.rds")
}
dim(hydrosprops.HYBRAS)
#> [1] 814 406.3.0.11 UNSODA
- Nemes, Attila; Schaap, Marcel; Leij, Feike J.; Wösten, J. Henk M. (2015). UNSODA 2.0: Unsaturated Soil Hydraulic Database. Database and program for indirect methods of estimating unsaturated hydraulic properties. US Salinity Laboratory - ARS - USDA. https://doi.org/10.15482/USDA.ADC/1173246. Accessed 2020-06-08.
if(!exists("hydrosprops.UNSODA")){
unsoda.LOC = read.csv("/mnt/diskstation/data/Soil_points/INT/UNSODA/general_c.csv")
#unsoda.LOC = unsoda.LOC[!unsoda.LOC$Lat==0,]
#plot(unsoda.LOC[,c("Long","Lat")])
unsoda.SOIL = read.csv("/mnt/diskstation/data/Soil_points/INT/UNSODA/soil_properties.csv")
#summary(unsoda.SOIL$k_sat)
## Soil water retention in lab:
tmp.hyd = read.csv("/mnt/diskstation/data/Soil_points/INT/UNSODA/lab_drying_h-t.csv")
#str(tmp.hyd)
tmp.hyd = tmp.hyd[!is.na(tmp.hyd$preshead),]
tmp.hyd$theta = tmp.hyd$theta*100
#head(tmp.hyd)
pr.lst = c(6,10,33,15000)
cl.lst = c("w6clod", "w10cld", "w3cld", "w15l2")
tmp.hyd.tbl = data.frame(code=unique(tmp.hyd$code), w6clod=NA, w10cld=NA, w3cld=NA, w15l2=NA)
for(i in 1:length(pr.lst)){
tmp.hyd.tbl[,cl.lst[i]] = plyr::join(tmp.hyd.tbl, tmp.hyd[which(tmp.hyd$preshead==pr.lst[i]),c("code","theta")], match="first")$theta
}
#head(tmp.hyd.tbl)
## ksat
kst.lev = read.csv("/mnt/diskstation/data/Soil_points/INT/UNSODA/comment_lab_sat_cond.csv", na.strings=c("","NA","No comment"))
kst.met = read.csv("/mnt/diskstation/data/Soil_points/INT/UNSODA/methodology.csv", na.strings=c("","NA","No comment"))
kst.met$comment_lsc = paste(plyr::join(kst.met[c("comment_lsc_ID")], kst.lev)$comment_lsc)
kst.met$comment_lsc[which(kst.met$comment_lsc=="NA")] = NA
kst.fld = read.csv("/mnt/diskstation/data/Soil_points/INT/UNSODA/comment_field_sat_cond.csv", na.strings=c("","NA","No comment"))
kst.met$comment_fsc = paste(plyr::join(kst.met[c("comment_fsc_ID")], kst.fld)$comment_fsc)
kst.met$comment_fsc[which(kst.met$comment_fsc=="NA")] = NA
summary(as.factor(kst.met$comment_lsc))
kst.met$comment_met = ifelse(is.na(kst.met$comment_lsc)&!is.na(kst.met$comment_fsc), paste("field", kst.met$comment_fsc), paste("lab", kst.met$comment_lsc))
unsoda.SOIL$comment_met = paste(plyr::join(unsoda.SOIL[c("code")], kst.met)$comment_met)
#summary(as.factor(unsoda.SOIL$comment_met))
sel.fld = unsoda.SOIL$comment_met %in% c("field Double ring infiltrometer","field Ponding", "field Steady infiltration")
unsoda.SOIL$ksat_lab[which(!sel.fld)] = unsoda.SOIL$k_sat[which(!sel.fld)]
unsoda.SOIL$ksat_field[is.na(unsoda.SOIL$ksat_lab)] = unsoda.SOIL$k_sat[is.na(unsoda.SOIL$ksat_lab)]
unsoda.col = join_all(list(unsoda.LOC, unsoda.SOIL, tmp.hyd.tbl))
#head(unsoda.col)
#summary(unsoda.col$OM_content)
unsoda.col$oc = signif(unsoda.col$OM_content/1.724, 4)
for(j in c("usiteid", "location_accuracy_min", "location_accuracy_max", "layer_sequence", "labsampnum", "db_13b", "COLEws", "w15bfm", "adod", "wrd_ws13", "cec7_cly", "w15cly", "cec_nh4", "ph_kcl", "wpg2")){ unsoda.col[,j] = NA }
unsoda.sel.h = c("code", "usiteid", "date", "Long", "Lat", "location_accuracy_min", "location_accuracy_max", "labsampnum", "layer_sequence", "depth_upper", "depth_lower", "horizon", "db_13b", "bulk_density", "COLEws", "w6clod", "w10cld", "w3cld", "w15l2", "w15bfm", "adod", "wrd_ws13", "cec7_cly", "w15cly", "Texture", "Clay", "Silt", "Sand", "oc", "ph_kcl", "pH", "CEC", "cec_nh4", "wpg2", "ksat_lab", "ksat_field")
hydrosprops.UNSODA = unsoda.col[,unsoda.sel.h]
hydrosprops.UNSODA$source_db = "UNSODA"
## corrected coordinates:
unsoda.ql = openxlsx::read.xlsx(xlsxFile, sheet = "UNSODA_degree")
hydrosprops.UNSODA$confidence_degree = plyr::join(hydrosprops.UNSODA["code"], unsoda.ql)$confidence_degree
hydrosprops.UNSODA$Texture = plyr::join(hydrosprops.UNSODA["code"], unsoda.ql)$tex_psda
hydrosprops.UNSODA$location_accuracy_min = plyr::join(hydrosprops.UNSODA["code"], unsoda.ql)$location_accuracy_min
hydrosprops.UNSODA$location_accuracy_max = plyr::join(hydrosprops.UNSODA["code"], unsoda.ql)$location_accuracy_max
## replace coordinates
unsoda.Long = plyr::join(hydrosprops.UNSODA["code"], unsoda.ql)$Improved_long
unsoda.Lat = plyr::join(hydrosprops.UNSODA["code"], unsoda.ql)$Improved_lat
hydrosprops.UNSODA$Long = ifelse(is.na(unsoda.Long), hydrosprops.UNSODA$Long, unsoda.Long)
hydrosprops.UNSODA$Lat = ifelse(is.na(unsoda.Long), hydrosprops.UNSODA$Lat, unsoda.Lat)
#hist(hydrosprops.UNSODA$w15l2, breaks=45, col="gray")
#hist(hydrosprops.UNSODA$ksat_lab, breaks=45, col="gray")
unsoda.rem = hydrosprops.UNSODA$code %in% unsoda.ql$code[is.na(unsoda.ql$additional_information)]
#summary(unsoda.rem)
hydrosprops.UNSODA = hydrosprops.UNSODA[unsoda.rem,]
## texture fractions sometimes need to be multiplied by 100!
#hydrosprops.UNSODA[hydrosprops.UNSODA$code==2220,]
sum.tex.1 = rowSums(hydrosprops.UNSODA[,c("Clay", "Silt", "Sand")], na.rm = TRUE)
sum.tex.r = which(sum.tex.1<1.2 & sum.tex.1>0)
for(j in c("Clay", "Silt", "Sand")){
hydrosprops.UNSODA[sum.tex.r,j] = hydrosprops.UNSODA[sum.tex.r,j] * 100
}
hydrosprops.UNSODA$project_url = "https://data.nal.usda.gov/dataset/unsoda-20-unsaturated-soil-hydraulic-database-database-and-program-indirect-methods-estimating-unsaturated-hydraulic-properties"
hydrosprops.UNSODA$citation_url = "https://doi.org/10.15482/USDA.ADC/1173246"
hydrosprops.UNSODA <- complete.vars(hydrosprops.UNSODA, coords = c("Long", "Lat"))
saveRDS.gz(hydrosprops.UNSODA, "/mnt/diskstation/data/Soil_points/INT/UNSODA/hydrosprops.UNSODA.rds")
}
dim(hydrosprops.UNSODA)
#> [1] 298 406.3.0.12 HYDROS
- Schindler, Uwe; Müller, Lothar (2015): Soil hydraulic functions of international soils measured with the Extended Evaporation Method (EEM) and the HYPROP device, Leibniz-Zentrum für Agrarlandschaftsforschung (ZALF) e.V.[doi: 10.4228/ZALF.2003.273]
if(!exists("hydrosprops.HYDROS")){
hydros.tbl = read.csv("/mnt/diskstation/data/Soil_points/INT/HydroS/int_rawret.csv", sep="\t", stringsAsFactors = FALSE, dec = ",")
hydros.tbl = hydros.tbl[!is.na(hydros.tbl$TENSION),]
#summary(hydros.tbl$TENSION)
hydros.tbl$TENSIONc = cut(hydros.tbl$TENSION, breaks=c(1,5,8,15,30,40,1000,15001))
#summary(hydros.tbl$TENSIONc)
hydros.tbl$WATER_CONTENT = hydros.tbl$WATER_CONTENT
#summary(hydros.tbl$WATER_CONTENT)
#head(hydros.tbl)
pr2.lst = c("(5,8]", "(8,15]","(30,40]","(1e+03,1.5e+04]")
cl.lst = c("w6clod", "w10cld", "w3cld", "w15l2")
hydros.tbl.df = data.frame(SITE_ID=unique(hydros.tbl$SITE_ID), w6clod=NA, w10cld=NA, w3cld=NA, w15l2=NA)
for(i in 1:length(pr2.lst)){
hydros.tbl.df[,cl.lst[i]] = plyr::join(hydros.tbl.df, hydros.tbl[which(hydros.tbl$TENSIONc==pr2.lst[i]),c("SITE_ID","WATER_CONTENT")], match="first")$WATER_CONTENT
}
#head(hydros.tbl.df)
## properties:
hydros.soil = read.csv("/mnt/diskstation/data/Soil_points/INT/HydroS/int_basicdata.csv", sep="\t", stringsAsFactors = FALSE, dec = ",")
#head(hydros.soil)
#plot(hydros.soil[,c("H","R")])
hydros.col = plyr::join(hydros.soil, hydros.tbl.df)
#summary(hydros.col$OMC)
hydros.col$oc = hydros.col$OMC/1.724
hydros.col$location_accuracy_min = 100
hydros.col$location_accuracy_max = NA
for(j in c("layer_sequence", "db_13b", "COLEws", "w15bfm", "adod", "wrd_ws13", "cec7_cly", "w15cly", "tex_psda", "clay_tot_psa", "silt_tot_psa", "sand_tot_psa", "oc", "ph_kcl", "ph_h2o", "cec_sum", "cec_nh4", "wpg2", "ksat_lab", "ksat_field")){ hydros.col[,j] = NA }
hydros.sel.h = c("SITE_ID", "SITE", "SAMP_DATE", "H", "R", "location_accuracy_min", "location_accuracy_max", "SAMP_NO", "layer_sequence", "TOP_DEPTH", "BOT_DEPTH", "HORIZON", "db_13b", "BULK_DENSITY", "COLEws", "w6clod", "w10cld", "w3cld", "w15l2", "w15bfm", "adod", "wrd_ws13", "cec7_cly", "w15cly", "tex_psda", "clay_tot_psa", "silt_tot_psa", "sand_tot_psa", "oc", "ph_kcl", "ph_h2o", "cec_sum", "cec_nh4", "wpg2", "ksat_lab", "ksat_field")
hydros.sel.h[which(!hydros.sel.h %in% names(hydros.col))]
hydrosprops.HYDROS = hydros.col[,hydros.sel.h]
hydrosprops.HYDROS$source_db = "HydroS"
hydrosprops.HYDROS$confidence_degree = 1
hydrosprops.HYDROS$project_url = "http://dx.doi.org/10.4228/ZALF.2003.273"
hydrosprops.HYDROS$citation_url = "https://doi.org/10.18174/odjar.v3i1.15763"
hydrosprops.HYDROS <- complete.vars(hydrosprops.HYDROS, coords = c("H","R"))
saveRDS.gz(hydrosprops.HYDROS, "/mnt/diskstation/data/Soil_points/INT/HYDROS/hydrosprops.HYDROS.rds")
}
dim(hydrosprops.HYDROS)
#> [1] 153 406.3.0.13 SWIG
- Rahmati, M., Weihermüller, L., Vanderborght, J., Pachepsky, Y. A., Mao, L., Sadeghi, S. H., … & Toth, B. (2018). Development and analysis of the Soil Water Infiltration Global database. Earth Syst. Sci. Data, 10, 1237–1263.
if(!exists("hydrosprops.SWIG")){
meta.tbl = read.csv("/mnt/diskstation/data/Soil_points/INT/SWIG/Metadata.csv", skip = 1, fill = TRUE, blank.lines.skip=TRUE, flush=TRUE, stringsAsFactors=FALSE)
swig.xy = read.table("/mnt/diskstation/data/Soil_points/INT/SWIG/Locations.csv", sep=";", dec = ",", stringsAsFactors=FALSE, header=TRUE, na.strings = c("-",""," "), fill = TRUE)
swig.xy$x = as.numeric(gsub(",", ".", swig.xy$x))
swig.xy$y = as.numeric(gsub(",", ".", swig.xy$y))
swig.xy = swig.xy[,1:8]
names(swig.xy)[3] = "EndDataset"
library(tidyr)
swig.xyf = tidyr::fill(swig.xy, c("Dataset","EndDataset"))
swig.xyf$N = swig.xyf$EndDataset - swig.xyf$Dataset + 1
swig.xyf$N = ifelse(swig.xyf$N<1,1,swig.xyf$N)
swig.xyf = swig.xyf[!is.na(swig.xyf$y),]
#plot(swig.xyf[,c("x","y")])
swig.xyf.df = swig.xyf[rep(seq_len(nrow(swig.xyf)), swig.xyf$N),]
rn = sapply(row.names(swig.xyf.df), function(i){as.numeric(strsplit(i, "\\.")[[1]][2])})
swig.xyf.df$Code = rowSums(data.frame(rn, swig.xyf.df$Dataset), na.rm = TRUE)
## bind together
swig.col = plyr::join(swig.xyf.df[,c("Code","x","y")], meta.tbl)
## aditional values for ksat
swig2.tbl = read.csv("/mnt/diskstation/data/Soil_points/INT/SWIG/Statistics.csv", fill = TRUE, blank.lines.skip=TRUE, sep=";", dec = ",", flush=TRUE, stringsAsFactors=FALSE)
#hist(log1p(as.numeric(swig2.tbl$Ks..cm.hr.)), breaks=45, col="gray")
swig.col$Ks..cm.hr. = as.numeric(plyr::join(swig.col["Code"], swig2.tbl[c("Code","Ks..cm.hr.")])$Ks..cm.hr.)
swig.col$Ks..cm.hr. = ifelse(swig.col$Ks..cm.hr. * 24 <= 0.01, NA, swig.col$Ks..cm.hr.)
swig.col$Ksat = ifelse(is.na(swig.col$Ksat), swig.col$Ks..cm.hr., swig.col$Ksat)
for(j in c("usiteid", "site_obsdate", "labsampnum", "layer_sequence", "hzn_desgn", "db_13b", "COLEws", "adod", "wrd_ws13", "w15bfm", "w15cly", "cec7_cly", "w6clod", "w10cld", "ph_kcl", "cec_nh4", "ksat_lab")){ swig.col[,j] = NA }
## depths are missing?
swig.col$hzn_top = 0
swig.col$hzn_bot = 20
swig.col$location_accuracy_min = NA
swig.col$location_accuracy_max = NA
swig.col$w15l2 = swig.col$PWP * 100
swig.col$w3cld = swig.col$FC * 100
swig.sel.h = c("Code", "usiteid", "site_obsdate", "x", "y", "location_accuracy_min", "location_accuracy_max", "labsampnum", "layer_sequence", "hzn_top", "hzn_bot", "hzn_desgn", "db_13b", "Db", "COLEws", "w6clod", "w10cld", "w3cld", "w15l2", "w15bfm", "adod", "wrd_ws13", "cec7_cly", "w15cly", "Texture.Class", "Clay", "Silt", "Sand", "OC", "ph_kcl", "pH", "CEC", "cec_nh4", "Gravel", "ksat_lab", "Ksat")
swig.sel.h[which(!swig.sel.h %in% names(swig.col))]
hydrosprops.SWIG = swig.col[,swig.sel.h]
hydrosprops.SWIG$source_db = "SWIG"
hydrosprops.SWIG$Ksat = hydrosprops.SWIG$Ksat * 24 ## convert to days
#hist(hydrosprops.SWIG$w3cld, breaks=45, col="gray")
#hist(log1p(hydrosprops.SWIG$Ksat), breaks=25, col="gray")
#summary(hydrosprops.SWIG$Ksat); summary(hydrosprops.UNSODA$ksat_lab)
## confidence degree
SWIG.ql = openxlsx::read.xlsx(xlsxFile, sheet = "SWIG_database_Confidence_degree")
hydrosprops.SWIG$confidence_degree = plyr::join(hydrosprops.SWIG["Code"], SWIG.ql)$confidence_degree
hydrosprops.SWIG$location_accuracy_min = plyr::join(hydrosprops.SWIG["Code"], SWIG.ql)$location_accuracy_min
hydrosprops.SWIG$location_accuracy_max = plyr::join(hydrosprops.SWIG["Code"], SWIG.ql)$location_accuracy_max
#summary(as.factor(hydrosprops.SWIG$confidence_degree))
## replace coordinates
SWIG.Long = plyr::join(hydrosprops.SWIG["Code"], SWIG.ql)$Improved_long
SWIG.Lat = plyr::join(hydrosprops.SWIG["Code"], SWIG.ql)$Improved_lat
hydrosprops.SWIG$x = ifelse(is.na(SWIG.Long), hydrosprops.SWIG$x, SWIG.Long)
hydrosprops.SWIG$y = ifelse(is.na(SWIG.Long), hydrosprops.SWIG$y, SWIG.Lat)
hydrosprops.SWIG$Texture.Class = plyr::join(hydrosprops.SWIG["Code"], SWIG.ql)$tex_psda
swig.lab = SWIG.ql$Code[which(SWIG.ql$Ksat_Method %in% c("Constant head method", "Constant Head Method", "Falling head method"))]
hydrosprops.SWIG$ksat_lab[hydrosprops.SWIG$Code %in% swig.lab] = hydrosprops.SWIG$Ksat[hydrosprops.SWIG$Code %in% swig.lab]
hydrosprops.SWIG$Ksat[hydrosprops.SWIG$Code %in% swig.lab] = NA
## remove duplicates
swig.rem = hydrosprops.SWIG$Code %in% SWIG.ql$Code[is.na(SWIG.ql$additional_information)]
#summary(swig.rem)
#Mode FALSE TRUE
#logical 200 6921
hydrosprops.SWIG = hydrosprops.SWIG[swig.rem,]
hydrosprops.SWIG = hydrosprops.SWIG[!duplicated(hydrosprops.SWIG$Code),]
## remove all ksat values < 0.01 ?
#summary(hydrosprops.SWIG$Ksat < 0.01)
hydrosprops.SWIG$project_url = "https://soil-modeling.org/resources-links/data-portal/swig"
hydrosprops.SWIG$citation_url = "https://doi.org/10.5194/essd-10-1237-2018"
hydrosprops.SWIG <- complete.vars(hydrosprops.SWIG, sel=c("w15l2","w3cld","ksat_lab","Ksat"), coords=c("x","y"))
saveRDS.gz(hydrosprops.SWIG, "/mnt/diskstation/data/Soil_points/INT/SWIG/hydrosprops.SWIG.rds")
}
dim(hydrosprops.SWIG)
#> [1] 3676 406.3.0.14 Pseudo-points
- Pseudo-observations using simulated points (world deserts)
if(!exists("hydrosprops.SIM")){
## 0 soil organic carbon + 98% sand content (deserts)
sprops.SIM = readRDS("/mnt/diskstation/data/LandGIS/training_points/soil_props/sprops.SIM.rds")
sprops.SIM$w10cld = 3.1
sprops.SIM$w3cld = 1.2
sprops.SIM$w15l2 = 0.8
sprops.SIM$tex_psda = "sand"
sprops.SIM$usiteid = sprops.SIM$lcv_admin0_fao.gaul_c_250m_s0..0cm_2015_v1.0
sprops.SIM$longitude_decimal_degrees = sprops.SIM$x
sprops.SIM$latitude_decimal_degrees = sprops.SIM$y
## Very approximate values for Ksat for shifting sand:
tax.r = raster::extract(raster("/mnt/diskstation/data/LandGIS/archive/predicted250m/sol_grtgroup_usda.soiltax_c_250m_s0..0cm_1950..2017_v0.1.tif"), sprops.SIM[,c("longitude_decimal_degrees","latitude_decimal_degrees")])
tax.leg = read.csv("/mnt/diskstation/data/LandGIS/archive/predicted250m/sol_grtgroup_usda.soiltax_c_250m_s0..0cm_1950..2017_v0.1.tif.csv")
tax.ksat_lab = aggregate(eth.tbl$ksat_lab, by=list(Group=eth.tbl$tax_grtgroup), FUN=mean, na.rm=TRUE)
tax.ksat_lab.sd = aggregate(eth.tbl$ksat_lab, by=list(Group=eth.tbl$tax_grtgroup), FUN=sd, na.rm=TRUE)
tax.ksat_field = aggregate(eth.tbl$ksat_field, by=list(Group=eth.tbl$tax_grtgroup), FUN=mean, na.rm=TRUE)
tax.leg$ksat_lab = join(tax.leg, tax.ksat_lab)$x
tax.leg$ksat_field = join(tax.leg, tax.ksat_field)$x
tax.sel = c("cryochrepts","cryorthods","torripsamments","haplustolls","torrifluvents")
sprops.SIM$ksat_field = join(data.frame(site_key=sprops.SIM$site_key, Number=tax.r), tax.leg[tax.leg$Group %in% tax.sel,])$ksat_field
sprops.SIM$ksat_lab = join(data.frame(site_key=sprops.SIM$site_key, Number=tax.r), tax.leg[tax.leg$Group %in% tax.sel,])$ksat_lab
#summary(sprops.SIM$ksat_lab)
#summary(sprops.SIM$ksat_field)
#View(sprops.SIM)
for(j in col.names[which(!col.names %in% names(sprops.SIM))]){ sprops.SIM[,j] <- NA }
sprops.SIM$project_url = "https://gitlab.com/openlandmap/global-layers"
sprops.SIM$citation_url = ""
hydrosprops.SIM = sprops.SIM[,col.names]
hydrosprops.SIM$confidence_degree = 30
saveRDS.gz(hydrosprops.SIM, "/mnt/diskstation/data/Soil_points/INT/hydrosprops.SIM.rds")
}
dim(hydrosprops.SIM)
#> [1] 8133 40
6.4  Bind all datasets
Bind all datasets
6.4.0.1 Bind and clean-up
Bind all tables / rename columns where necessary:
ls(pattern=glob2rx("hydrosprops.*"))
#> [1] "hydrosprops.AfSPDB" "hydrosprops.EGRPR" "hydrosprops.ETH"
#> [4] "hydrosprops.FRED" "hydrosprops.HYBRAS" "hydrosprops.HYDROS"
#> [7] "hydrosprops.ISIS" "hydrosprops.NCSS" "hydrosprops.NPDB"
#> [10] "hydrosprops.SIM" "hydrosprops.SPADE2" "hydrosprops.SWIG"
#> [13] "hydrosprops.UNSODA" "hydrosprops.WISE"
tot_sprops = dplyr::bind_rows(lapply(ls(pattern=glob2rx("hydrosprops.*")), function(i){ mutate_all(setNames(get(i), col.names), as.character) }))
## convert to numeric:
for(j in c("longitude_decimal_degrees", "latitude_decimal_degrees", "location_accuracy_min", "location_accuracy_max", "layer_sequence", "hzn_top","hzn_bot", "oc", "ph_h2o", "ph_kcl", "db_od", "clay_tot_psa", "sand_tot_psa","silt_tot_psa", "wpg2", "db_13b", "COLEws", "w15cly", "w6clod", "w10cld", "w3cld", "w15l2", "w15bfm", "adod", "wrd_ws13","cec7_cly", "cec_sum", "cec_nh4", "ksat_lab","ksat_field")){
tot_sprops[,j] = as.numeric(tot_sprops[,j])
}
#> Warning: NAs introduced by coercion
#> Warning: NAs introduced by coercion
#> Warning: NAs introduced by coercion
#> Warning: NAs introduced by coercion
#> Warning: NAs introduced by coercion
#head(tot_sprops)
## rename some columns:
tot_sprops = plyr::rename(tot_sprops, replace = c("db_od" = "db", "ph_h2o" = "ph_h2o_v", "oc" = "oc_v"))
summary(as.factor(tot_sprops$source_db))
#> AfSPDB Australian_ksat_data Belgian_ksat_data
#> 10720 118 145
#> Canada_NPDB China_ksat_data ETH_literature
#> 404 209 1954
#> Florida_ksat_data FRED HYBRAS
#> 6532 3761 814
#> HydroS ISRIC_ISIS ISRIC_WISE
#> 153 1176 1325
#> Russia_EGRPR SIMULATED SPADE2
#> 1138 8133 1182
#> SWIG Tibetan_plateau_ksat_data UNSODA
#> 3676 65 298
#> USDA_NCSS
#> 113991Add unique row identifier
tot_sprops$uuid = uuid::UUIDgenerate(use.time=TRUE, n=nrow(tot_sprops))and unique location based on the Open Location Code:
tot_sprops$olc_id = olctools::encode_olc(tot_sprops$latitude_decimal_degrees, tot_sprops$longitude_decimal_degrees, 11)
length(levels(as.factor(tot_sprops$olc_id)))
#> [1] 250756.4.0.2 Quality-control spatial locations
Unique locations:
tot_sprops.pnts = tot_sprops[!duplicated(tot_sprops$olc_id),c("site_key", "source_db", "longitude_decimal_degrees", "latitude_decimal_degrees", "site_obsdate", "olc_id", "project_url", "citation_url")]
coordinates(tot_sprops.pnts) <- ~ longitude_decimal_degrees + latitude_decimal_degrees
proj4string(tot_sprops.pnts) <- "+init=epsg:4326"Remove points falling in the sea or similar:
if(!exists("ov.sprops")){
#mask = terra::rast("./layers1km/lcv_landmask_esacci.lc.l4_c_1km_s0..0cm_2000..2015_v1.0.tif")
mask = terra::rast("/mnt/diskstation/data/LandGIS/layers250m/lcv_landmask_esacci.lc.l4_c_250m_s0..0cm_2000..2015_v1.0.tif")
ov.sprops <- terra::extract(mask, terra::vect(tot_sprops.pnts))
summary(as.factor(ov.sprops[,2]))
if(sum(is.na(ov.sprops[,2]))>0 | sum(ov.sprops[,2]==2)>0){
rem.lst = which(is.na(ov.sprops[,2]) | ov.sprops[,2]==2 | ov.sprops[,2]==4)
rem.sp = tot_sprops.pnts$site_key[rem.lst]
tot_sprops.pnts = tot_sprops.pnts[-rem.lst,]
} else {
rem.sp = NA
}
}
## final number of unique spatial locations:
nrow(tot_sprops.pnts)
#> [1] 250756.4.0.3 Clean-up
Clean up typos and physically impossible values:
for(j in c("clay_tot_psa", "sand_tot_psa", "silt_tot_psa", "wpg2", "w6clod", "w10cld", "w3cld", "w15l2")){
tot_sprops[,j] = ifelse(tot_sprops[,j]>100|tot_sprops[,j]<0, NA, tot_sprops[,j])
}
for(j in c("ph_h2o_v","ph_kcl")){
tot_sprops[,j] = ifelse(tot_sprops[,j]>12|tot_sprops[,j]<2, NA, tot_sprops[,j])
}
#hist(tot_sprops$db_od)
for(j in c("db")){
tot_sprops[,j] = ifelse(tot_sprops[,j]>2.4|tot_sprops[,j]<0.05, NA, tot_sprops[,j])
}
#summary(tot_sprops$ksat_lab)
for(j in c("ksat_lab","ksat_field")){
tot_sprops[,j] = ifelse(tot_sprops[,j] <=0, NA, tot_sprops[,j])
}
#hist(tot_sprops$oc)
for(j in c("oc_v")){
tot_sprops[,j] = ifelse(tot_sprops[,j]>90|tot_sprops[,j]<0, NA, tot_sprops[,j])
}
tot_sprops$hzn_depth = tot_sprops$hzn_top + (tot_sprops$hzn_bot-tot_sprops$hzn_top)/2
#tot_sprops = tot_sprops[!is.na(tot_sprops$hzn_depth),]
## texture fractions check:
sum.tex.T = rowSums(tot_sprops[,c("clay_tot_psa", "silt_tot_psa", "sand_tot_psa")], na.rm = TRUE)
which(sum.tex.T<1.2 & sum.tex.T>0)
#> [1] 9334 9979 12371 22431 22441 81311 81312 81313 93971 150217
for(i in which(sum.tex.T<1.2 & sum.tex.T>0)){
for(j in c("clay_tot_psa", "silt_tot_psa", "sand_tot_psa")){
tot_sprops[i,j] <- NA
}
}6.4.0.4 Histogram plots
library(ggplot2)
#ggplot(tot_sprops[tot_sprops$w15l2<100,], aes(x=source_db, y=w15l2)) + geom_boxplot() + theme(axis.text.x = element_text(angle = 90, hjust = 1))
ggplot(tot_sprops[tot_sprops$w3cld<100,], aes(x=source_db, y=w3cld)) + geom_boxplot() + theme(axis.text.x = element_text(angle = 90, hjust = 1))
#> Warning: Removed 68935 rows containing non-finite values (stat_boxplot).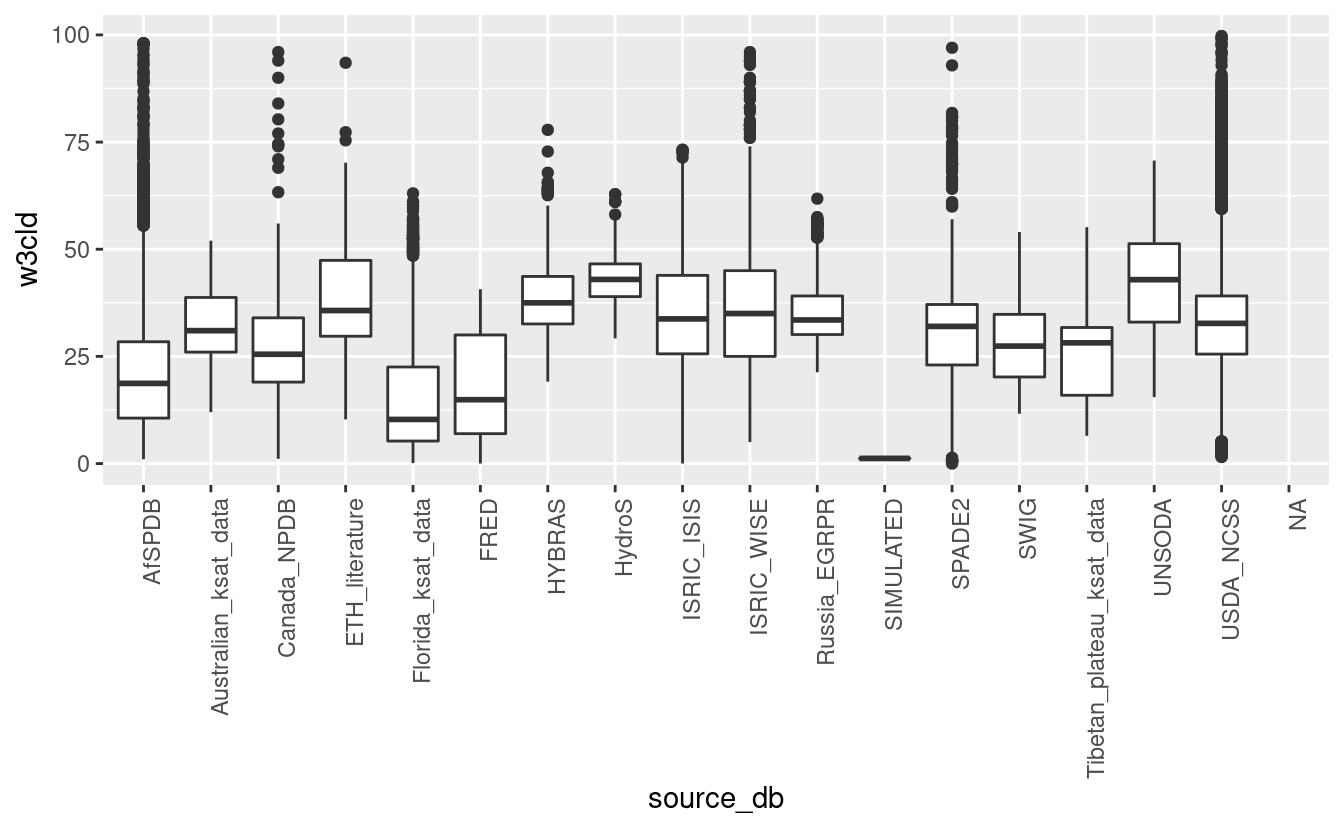
ggplot(tot_sprops, aes(x=source_db, y=db)) + geom_boxplot() + theme(axis.text.x = element_text(angle = 90, hjust = 1))
#> Warning: Removed 69588 rows containing non-finite values (stat_boxplot).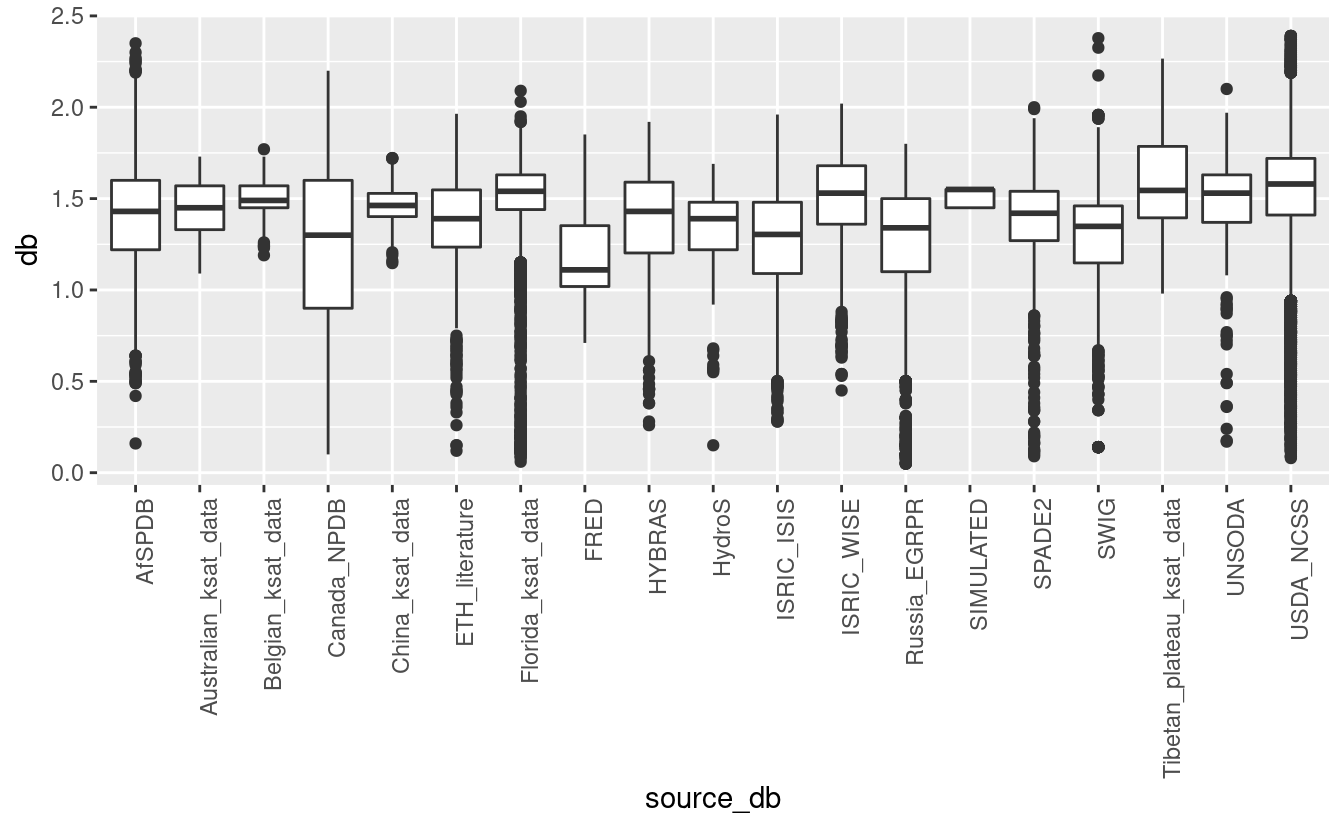
ggplot(tot_sprops, aes(x=source_db, y=ph_h2o_v)) + geom_boxplot() + theme(axis.text.x = element_text(angle = 90, hjust = 1))
#> Warning: Removed 122423 rows containing non-finite values (stat_boxplot).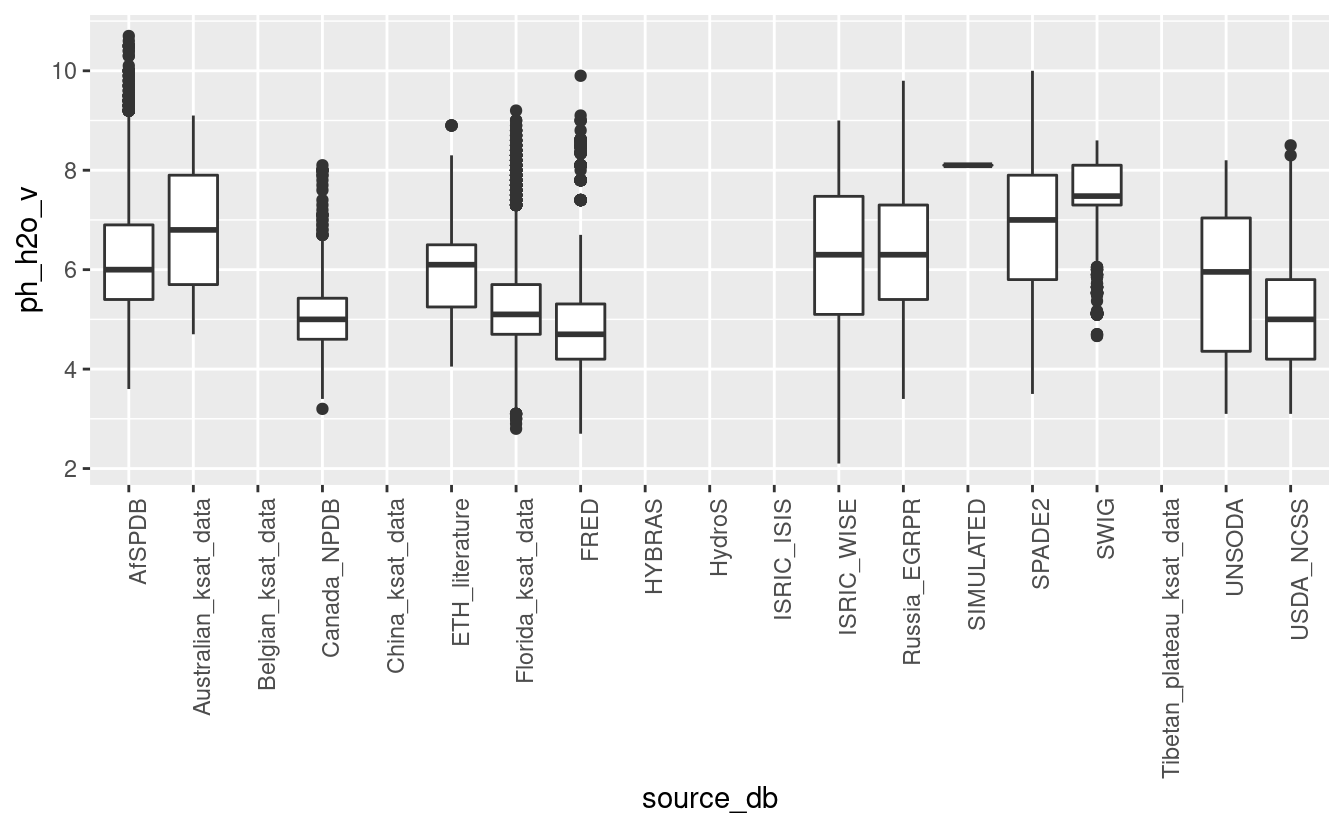
ggplot(tot_sprops, aes(x=source_db, y=log10(ksat_field+1))) + geom_boxplot() + theme(axis.text.x = element_text(angle = 90, hjust = 1))
#> Warning: Removed 144683 rows containing non-finite values (stat_boxplot).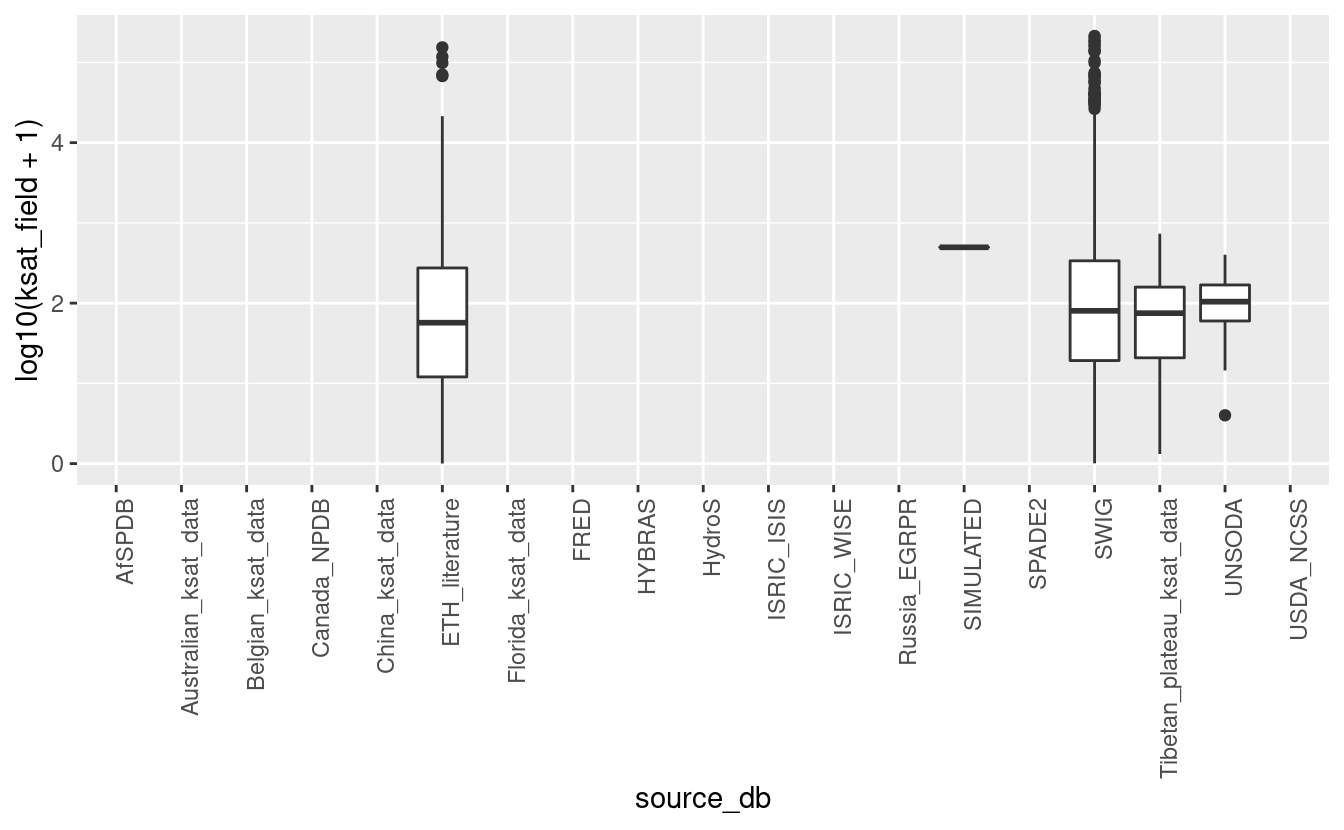
ggplot(tot_sprops, aes(x=source_db, y=log1p(ksat_lab))) + geom_boxplot() + theme(axis.text.x = element_text(angle = 90, hjust = 1))
#> Warning: Removed 139595 rows containing non-finite values (stat_boxplot).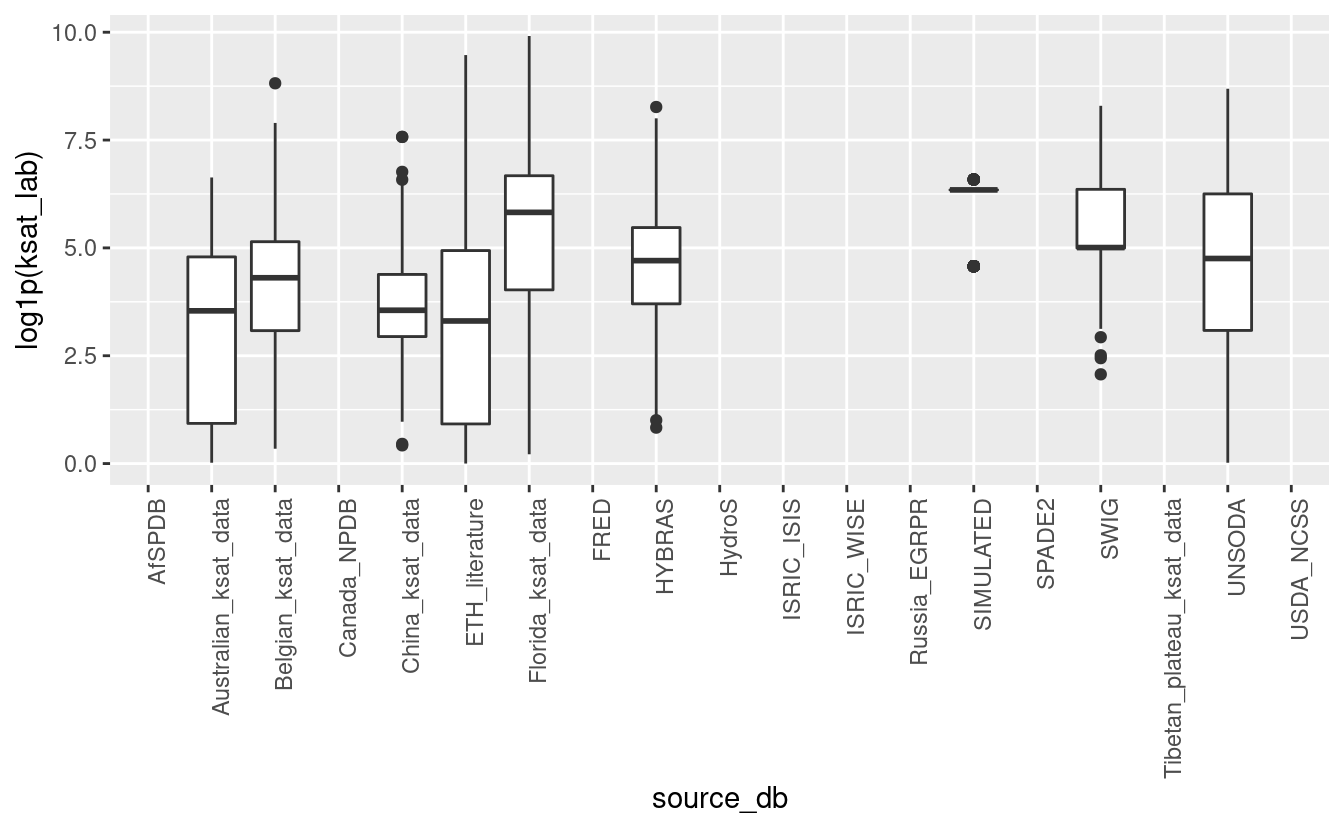
6.4.0.5 Convert to wide format
Add layer_sequence where missing since this is needed to be able to convert to wide
format:
#summary(tot_sprops$layer_sequence)
tot_sprops$dsiteid = paste(tot_sprops$source_db, tot_sprops$site_key, tot_sprops$site_obsdate, sep="_")
if(!exists("l.s1")){
library(dplyr)
## Note: takes >1 min
l.s1 <- tot_sprops[,c("olc_id","hzn_depth")] %>% group_by(olc_id) %>% mutate(layer_sequence.f = data.table::frank(hzn_depth, ties.method = "first"))
tot_sprops$layer_sequence.f = ifelse(is.na(tot_sprops$layer_sequence), l.s1$layer_sequence.f, tot_sprops$layer_sequence)
tot_sprops$layer_sequence.f = ifelse(tot_sprops$layer_sequence.f>6, 6, tot_sprops$layer_sequence.f)
}Convert long table to wide table format so that each depth gets unique column:
if(!exists("tot_sprops.w")){
library(data.table)
hor.names.s = c("hzn_top", "hzn_bot", "hzn_desgn", "db", "w6clod", "w3cld", "w15l2", "adod", "wrd_ws13", "tex_psda", "clay_tot_psa", "silt_tot_psa", "sand_tot_psa", "oc_v", "ph_kcl", "ph_h2o_v", "cec_sum", "cec_nh4", "wpg2", "ksat_lab", "ksat_field", "uuid")
tot_sprops.w = data.table::dcast( as.data.table(tot_sprops),
formula = olc_id ~ layer_sequence.f,
value.var = hor.names.s,
fun=function(x){ x[1] },
verbose = FALSE)
}
tot_sprops_w.pnts = tot_sprops.pnts
tot_sprops_w.pnts@data = plyr::join(tot_sprops.pnts@data, tot_sprops.w)
#> Joining by: olc_idWrite all soil profiles using a wide format:
6.4.0.6 Ksat dataset:
sel.compl = !is.na(tot_sprops$longitude_decimal_degrees) & !is.na(tot_sprops$w3cld) & !is.na(tot_sprops$ph_h2o_v) & !is.na(tot_sprops$clay_tot_psa) & !is.na(tot_sprops$oc_v)
summary(sel.compl)
#> Mode FALSE TRUE
#> logical 131042 24752
## complete ksat_field points
tot_sprops.pnts.C = tot_sprops[!is.na(tot_sprops$longitude_decimal_degrees) & !is.na(tot_sprops$latitude_decimal_degrees) & (!is.na(tot_sprops$ksat_lab) | !is.na(tot_sprops$ksat_field)) & !tot_sprops$source_db == "SIMULATED",]
sum.NA = sapply(tot_sprops.pnts.C, function(i){sum(is.na(i))})
tot_sprops.pnts.C = tot_sprops.pnts.C[,!sum.NA==nrow(tot_sprops.pnts.C)]
tot_sprops.pnts.C$w10cld = NULL
tot_sprops.pnts.C$w6clod = NULL
tot_sprops.pnts.C$w15bfm = NULL
tot_sprops.pnts.C$site_obsdate = NULL
tot_sprops.pnts.C$confidence_degree = NULL
tot_sprops.pnts.C$cec_sum = NULL
tot_sprops.pnts.C$wpg2 = NULL
tot_sprops.pnts.C$uuid = NULL
dim(tot_sprops.pnts.C)
#> [1] 13258 256.4.0.7 RDS files
Plot in Goode Homolozine projection and save final objects:
if(!file.exists("./img/sol_hydro.pnts_sites.png")){
tot_sprops.pnts_sf <- st_as_sf(tot_sprops.pnts[1])
plot_gh(tot_sprops.pnts_sf, out.pdf="./img/sol_hydro.pnts_sites.pdf")
system("pdftoppm ./img/sol_hydro.pnts_sites.pdf ./img/sol_hydro.pnts_sites -png -f 1 -singlefile")
system("convert -crop 1280x575+36+114 ./img/sol_hydro.pnts_sites.png ./img/sol_hydro.pnts_sites.png")
}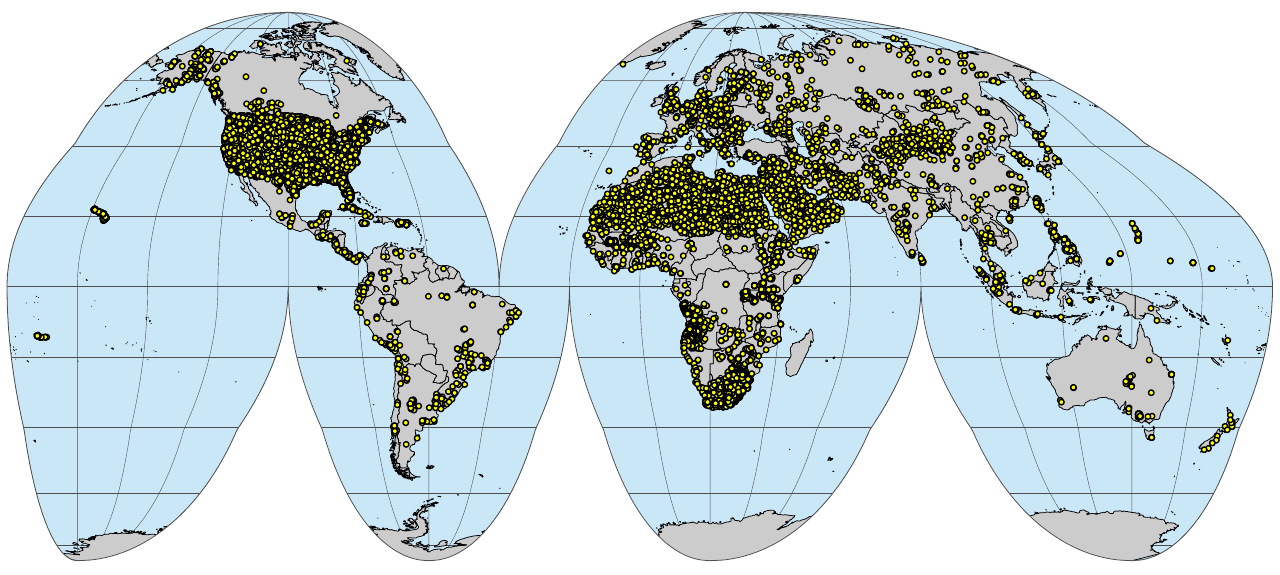
(#fig:sol_hydro.pnts_sites)Soil profiles and soil samples with physical and hydraulic soil properties properties global compilation.
Fig. 1: Soil profiles and soil samples with physical and hydraulic soil properties properties global compilation.
if(!file.exists("./img/sol_ksat.pnts_sites.png")){
sel.ks = which(tot_sprops.pnts$location_id %in% tot_sprops.pnts.C$location_id)
tot_spropsC.pnts_sf <- st_as_sf(tot_sprops.pnts[sel.ks, 1])
plot_gh(tot_spropsC.pnts_sf, out.pdf="./img/sol_ksat.pnts_sites.pdf")
system("pdftoppm ./img/sol_ksat.pnts_sites.pdf ./img/sol_ksat.pnts_sites -png -f 1 -singlefile")
system("convert -crop 1280x575+36+114 ./img/sol_ksat.pnts_sites.png ./img/sol_ksat.pnts_sites.png")
}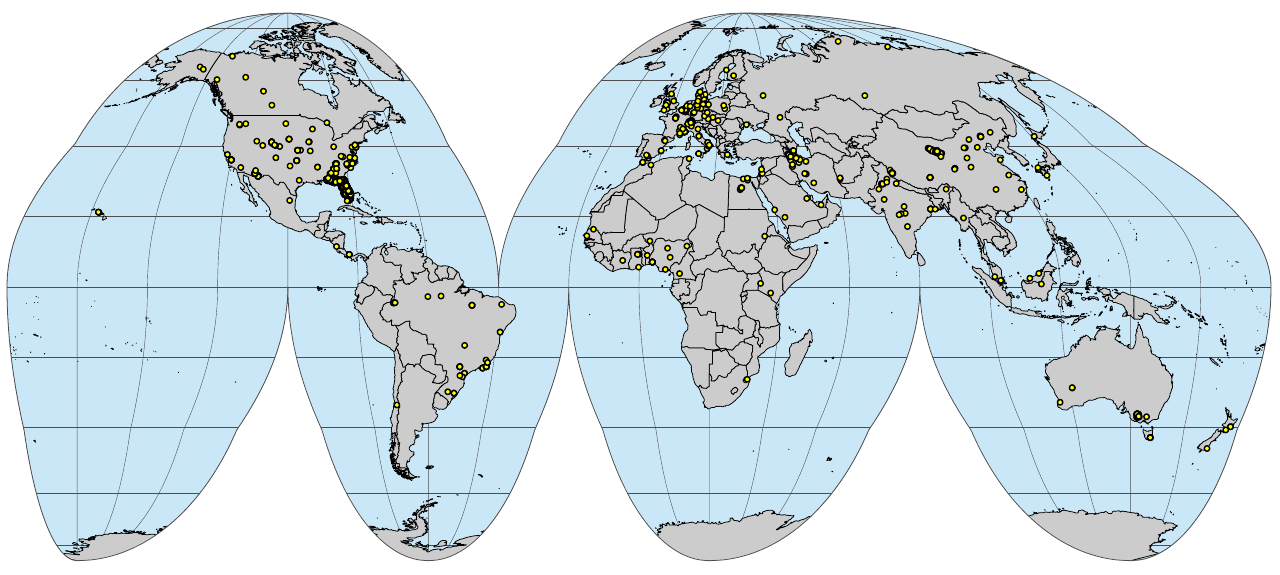
(#fig:sol_ksat.pnts_sites)Soil profiles and soil samples with Ksat measurements global compilation
Fig. 2: Soil profiles and soil samples with Ksat measurements global compilation.
6.5  Overlay www.OpenLandMap.org layers
Overlay www.OpenLandMap.org layers
Load the tiling system (1 degree grid representing global land mask) and run spatial overlay in parallel:
if(!exists("rm.sol")){
tile.pol = readOGR("./tiles/global_tiling_100km_grid.gpkg")
#length(tile.pol)
ov.sol <- extract.tiled(obj=tot_sprops.pnts, tile.pol=tile.pol, path="/data/tt/LandGIS/grid250m", ID="ID", cpus=64)
## Valid predictors:
pr.vars = unique(unlist(sapply(c("fapar", "landsat", "lc100", "mod09a1", "mod11a2", "alos.palsar", "sm2rain", "irradiation_solar.atlas", "usgs.ecotapestry", "floodmap.500y", "bioclim", "water.table.depth_deltares", "snow.prob_esacci", "water.vapor_nasa.eo", "wind.speed_terraclimate", "merit.dem_m", "merit.hydro_m", "cloud.fraction_earthenv", "water.occurance_jrc", "wetlands.cw_upmc", "pb2002"), function(i){names(ov.sol)[grep(i, names(ov.sol))]})))
str(pr.vars)
## 349
#saveRDS.gz(ov.sol, "/mnt/diskstation/data/Soil_points/ov.sol_hydro.pnts_horizons.rds")
#ov.sol <- readRDS.gz("/mnt/diskstation/data/Soil_points/ov.sol_hydro.pnts_horizons.rds")
## Final regression matrix:
rm.sol = plyr::join(tot_sprops, ov.sol[,c("olc_id", pr.vars)])
## check that there are no duplicates
sum(duplicated(rm.sol$uuid))
rm.ksat = rm.sol[(!is.na(rm.sol$ksat_lab) | !is.na(rm.sol$ksat_field)),]
}
dim(rm.sol)Save final analysis-ready objects:
saveRDS.gz(tot_sprops, "./out/rds/sol_hydro.pnts_horizons.rds")
saveRDS.gz(tot_sprops.pnts, "/mnt/diskstation/data/Soil_points/sol_hydro.pnts_sites.rds")
## reorder columns
tot_sprops.pnts.C$ID = 1:nrow(tot_sprops.pnts.C)
tot_sprops.pnts.C = tot_sprops.pnts.C[,c(which(names(tot_sprops.pnts.C)=="ID"), which(!names(tot_sprops.pnts.C)=="ID"))]
saveRDS.gz(tot_sprops.pnts.C, "./out/rds/sol_ksat.pnts_horizons.rds")
#library(farff)
#writeARFF(tot_sprops, "./out/arff/sol_hydro.pnts_horizons.arff", overwrite = TRUE)
#writeARFF(tot_sprops.pnts.C, "./out/arff/sol_ksat.pnts_horizons.arff", overwrite = TRUE)
## compressed CSV
write.csv(tot_sprops, file=gzfile("./out/csv/sol_hydro.pnts_horizons.csv.gz"))
write.csv(tot_sprops.pnts.C, file=gzfile("./out/csv/sol_ksat.pnts_horizons.csv.gz"), row.names = FALSE)
saveRDS.gz(rm.sol, "./out/rds/sol_hydro.pnts_horizons_rm.rds")
saveRDS.gz(rm.ksat, "./out/rds/sol_ksat.pnts_horizons_rm.rds")Save temp object:
#rm(rm.sol); gc()
save.image.pigz(file="soilhydro.RData")
## rmarkdown::render("Index.rmd")